bioMONAI

Overview
bioMONAI is a low-code Python-based platform for developing and deploying deep learning models in biomedical imaging built on top of the MONAI framework, fastai, and TorchIO. This project aims to facilitate interoperability, reproducibility, and community collaboration in biomedical research.
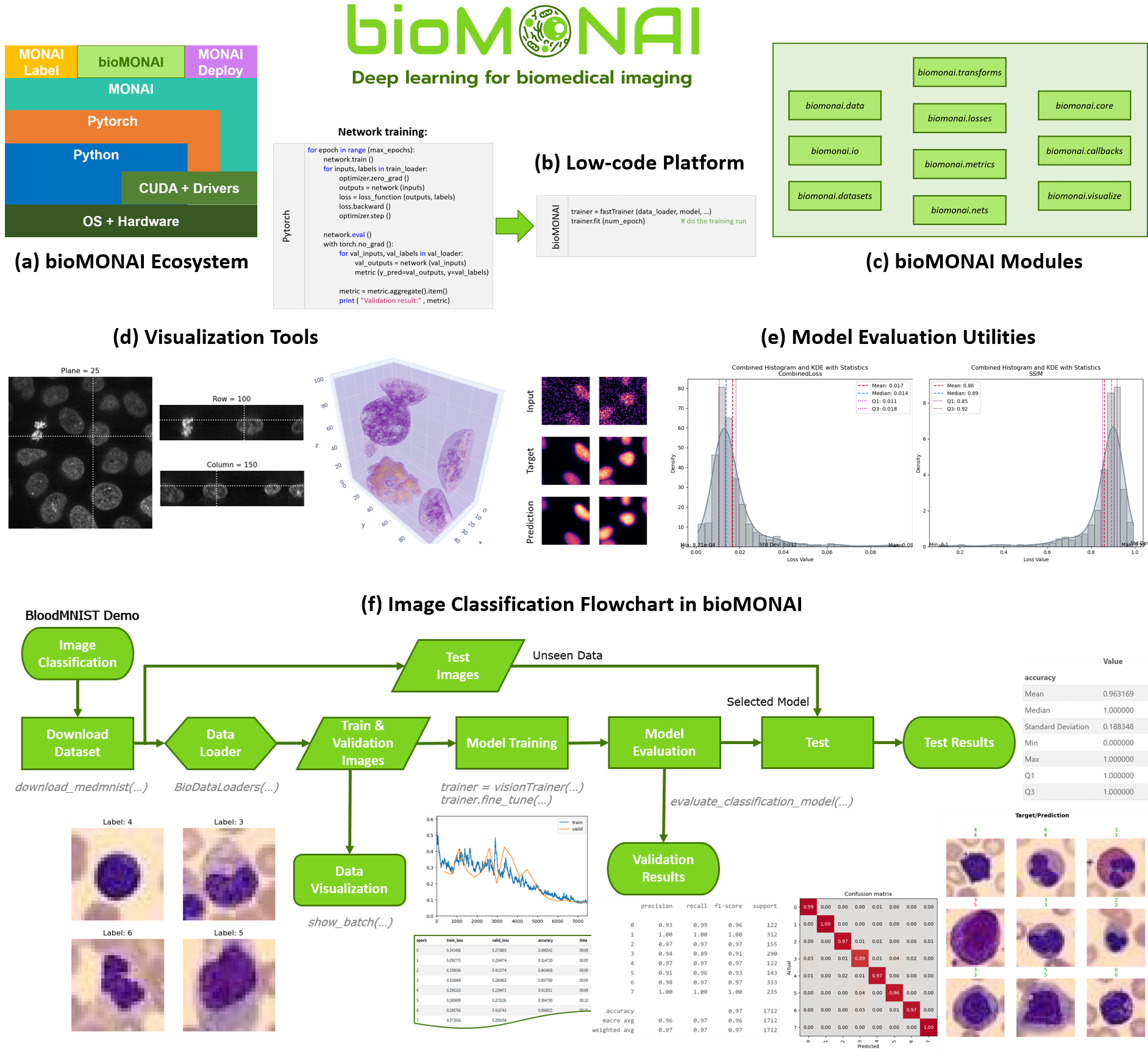
Table of Contents
Installation
Before starting, ensure you have Conda installed (Anaconda or Miniconda) on your system.
For Windows users, it is highly recommended to install and run BioMONAI within WSL2 (Windows Subsystem for Linux 2) for optimal compatibility and performance. Additionally, ensure that the NVIDIA CUDA toolkit is properly installed inside WSL2 and that the nvcc compiler is available in your environment (nvcc --version should return the installed version).
Installation Steps
Clone the repository:
git clone https://github.com/deepclem/biomonai.git cd biomonaiCreate a new Conda environment and install dependencies:
conda env create -f bioMONAI-linux.ymlActivate the environment and install BioMONAI in editable mode:
conda activate bioMONAI-env pip install -e .
Notes:
If you are not using WSL2 on Windows, some functionalities may not work as expected.
Make sure your system’s CUDA installation is compatible with PyTorch and MONAI.
You can verify CUDA availability with:
nvcc --versionReplace
bioMONAI-linux.ymlwith a different environment file if you’re using another OS or configuration.
Getting Started
To get started with bioMONAI, we recommend exploring our tutorials, which guide you through model training for various tasks such as classification and denoising.
| Notebook | Open in Colab |
|---|---|
| Tutorial: Classification 2D This notebook provides a comprehensive guide on training deep learning models for 2D image classification tasks, covering data loading, preprocessing, model building, training, and evaluation. |
|
| Tutorial: Denoising 2D This notebook offers a detailed guide on applying deep learning techniques to denoise biological microscopy images. It covers data preparation, model architecture, training processes, and evaluation methods, providing a comprehensive resource for enhancing image quality in biological research. |
Usage
To use bioMONAI for your own projects, follow these steps:
Create a new Jupyter notebook or open an existing one.
Import necessary modules:
import bioMONAIStart coding! You can now leverage MONAI’s capabilities alongside the interactive features of Jupyter notebooks.
Contributing
We welcome contributions from the community! To contribute to BioMONAI nbs, follow these steps:
Fork the repository on GitHub.
Clone your fork:
git clone https://github.com/your_username/biomonai.git cd biomonaiCreate a new Conda environment and install dependencies:
conda env create --file bioMONAI-env.ymlActivate the environment and install MONAI in dev mode:
conda activate bioMONAI-env pip install -e .[dev]Create a new branch for your changes:
git checkout -b feature/new-featureMake your changes and commit them:
git add . git commit -m "Add new feature: <feature description>"Push to your fork and create a pull request on GitHub.
Wait for the review, and merge if everything looks good!
System Requirements
BioMONAI is built on top of MONAI and fastai, leveraging their core functionalities while adding features to simplify and enhance the use of AI models in biomedical applications. Despite these additional features, BioMONAI does not introduce extra computational overhead beyond what is required by the underlying frameworks.
As such, the system requirements are aligned with those recommended by MONAI:
RAM: Minimum 32 GB
Storage: Minimum 50 GB of available SSD space
GPU: Highly recommended for efficient training and inference
Compatible with NVIDIA GPUs supporting CUDA
At least 8 GB of GPU memory recommended
CPU: Supported, but significantly slower execution compared to GPU
Operating System: Linux (Ubuntu 20.04+ recommended), macOS, or Windows 10/11
⚠️ Note: While CPU-only execution is possible, users should expect substantial increases in training and inference time without a GPU.
License
bioMONAI is released under the Apache 2.0 license. See LICENSE for more details.
Contact
If you have any questions or need further assistance, please open an issue on GitHub or contact us directly at:
- Project Lead: Biagio Mandracchia
- Contributors: Sara Cruz-Adrados, Juan Pita-López, Rosa-María Menchón-Lara, Miguel Ángel Martín Fernandez



