from bioMONAI.data import *
from bioMONAI.transforms import *
from bioMONAI.core import *
from bioMONAI.core import Path
from bioMONAI.losses import *
from bioMONAI.metrics import *
from bioMONAI.datasets import download_medmnist
from bioMONAI.visualize import show_images_grid, mosaic_image_3d
from bioMONAI.data import get_image_filesImage Classification 3D
Setup imports
device = get_device()
print(device)cudaDownload and store the dataset
These lines of code will download the SynapseMNIST3D dataset and set up the paths for training, validation, and test datasets.
The download_medmnist function is used to download the dataset, and the paths are organized to easily access different parts of the dataset for training, validation, and testing purposes. The data_path is updated to point to the ‘synapsemnist3d’ directory, and then separate paths are created for the ‘train’, ‘val’, and ‘test’ subdirectories. This organization helps in easily accessing the different parts of the dataset for training, validation, and testing purposes.
You can customize the data_flag to download different datasets available in the MedMNIST collection. Additionally, you can modify the data_path to change the location where the dataset is stored. If you have a specific directory structure in mind, ensure that the paths for train_path, val_path, and test_path correctly reflect your desired organization. This flexibility allows you to adapt the code to various datasets and storage requirements.
data_flag = 'synapsemnist3d'
data_path = Path('../_data/medmnist_data/')
info = download_medmnist(data_flag, data_path, download_only=True)
data_path = data_path/'synapsemnist3d'
train_path = data_path/'train'
val_path = data_path/'val'
test_path = data_path/'test'Downloading https://zenodo.org/records/10519652/files/synapsemnist3d.npz?download=1 to ../_data/medmnist_data/synapsemnist3d/synapsemnist3d.npz100%|██████████| 38034583/38034583 [00:10<00:00, 3689266.37it/s]Using downloaded and verified file: ../_data/medmnist_data/synapsemnist3d/synapsemnist3d.npz
Using downloaded and verified file: ../_data/medmnist_data/synapsemnist3d/synapsemnist3d.npz
Saving training images to ../_data/medmnist_data/synapsemnist3d...100%|██████████| 1230/1230 [00:07<00:00, 160.10it/s]Saving validation images to ../_data/medmnist_data/synapsemnist3d...100%|██████████| 177/177 [00:01<00:00, 144.49it/s]Saving test images to ../_data/medmnist_data/synapsemnist3d...100%|██████████| 352/352 [00:02<00:00, 163.47it/s]Removed synapsemnist3d.npz
Datasets downloaded to ../_data/medmnist_data/synapsemnist3d
Dataset info for 'synapsemnist3d': {'python_class': 'SynapseMNIST3D', 'description': 'The SynapseMNIST3D is a new 3D volume dataset to classify whether a synapse is excitatory or inhibitory. It uses a 3D image volume of an adult rat acquired by a multi-beam scanning electron microscope. The original data is of the size 100×100×100um^3 and the resolution 8×8×30nm^3, where a (30um)^3 sub-volume was used in the MitoEM dataset with dense 3D mitochondria instance segmentation labels. Three neuroscience experts segment a pyramidal neuron within the whole volume and proofread all the synapses on this neuron with excitatory/inhibitory labels. For each labeled synaptic location, we crop a 3D volume of 1024×1024×1024nm^3 and resize it into 28×28×28 voxels. Finally, the dataset is randomly split with a ratio of 7:1:2 into training, validation and test set.', 'url': 'https://zenodo.org/records/10519652/files/synapsemnist3d.npz?download=1', 'MD5': '1235b78a3cd6280881dd7850a78eadb6', 'url_64': 'https://zenodo.org/records/10519652/files/synapsemnist3d_64.npz?download=1', 'MD5_64': '43bd14ebf3af9d3dd072446fedc14d5e', 'task': 'binary-class', 'label': {'0': 'inhibitory synapse', '1': 'excitatory synapse'}, 'n_channels': 1, 'n_samples': {'train': 1230, 'val': 177, 'test': 352}, 'license': 'CC BY 4.0'}Create Dataloader
Customize DataLoader
In the next cell, we will create a DataLoader for the SynapseMNIST3D dataset.
The BioDataLoaders.class_from_folder() method is used to load the dataset from the specified paths and apply transformations to the images. This method is designed for classification tasks and datasets organized in folders. Indeed, it assumes that the images are organized in subfolders whose names represents the image labels.
We will set the batch size to 8 and apply the following transformations: - ScaleIntensity(): Scales the intensity of the images. - RandRot90(prob=0.5, spatial_axes=(1,2)): Randomly rotates the images by 90 degrees with a probability of 0.5 along the specified spatial axes. - Resize(32): Resizes the images to 32x32x32.
You can customize the DataLoader by changing the batch size, adding or removing transformations, or modifying the paths to the dataset. For example, you can increase the batch size for faster training or add more complex transformations to augment the dataset.
The show_summary parameter is set to True to display a summary of the dataset and transformations applied.
After creating the DataLoader, we will print the number of training and validation images to verify that the dataset has been loaded correctly.
Parameters:
data_path: The root directory where the dataset is stored.train: The subdirectory containing the training images.valid: The subdirectory containing the validation images.vocab: The vocabulary or labels for the dataset.item_tfms: A list of transformations to apply to each image individually. Examples includeScaleIntensity(),RandRot90(), andResize().batch_tfms: A list of transformations to apply to a batch of images. This can be set toNoneif no batch transformations are needed.img_cls: The class to use for loading images. For 3D images, this is typicallyBioImageStack.bs: The batch size, which determines how many images are processed together in each batch.show_summary: A boolean flag to display a summary of the dataset and the transformations applied.
Example Usage:
batch_size = 4
data_ops = {
'train': 'train', # folder for training data
'valid': 'val', # folder for validation data
'vocab': info['label'], # list of class labels
'img_cls': BioImageStack, # class to use for images
'bs': batch_size, # batch size
'batch_tfms': None,
'item_tfms': [ScaleIntensity(),
RandRot90(prob=0.75, spatial_axes=(1,2)),
Resize(32)],
}
data = BioDataLoaders.class_from_folder(
data_path, # root directory for data
show_summary=False, # print summary of the data
**data_ops, # rest of the method arguments
)
# print length of training and validation datasets
print('train images:', len(data.train_ds.items), '\nvalidation images:', len(data.valid_ds.items))train images: 1230
validation images: 177Display a Batch of Images
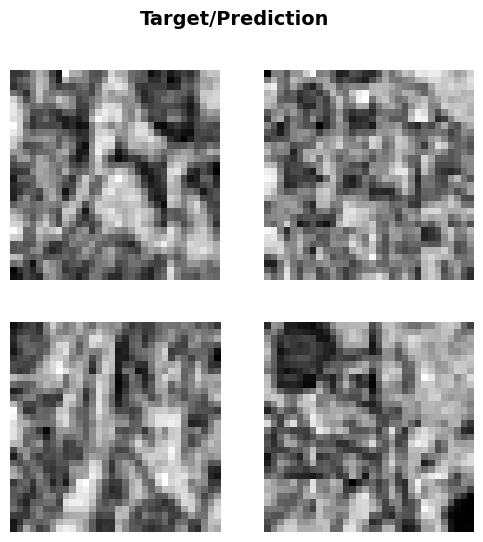
In the next cell, we will display a batch of images from the training dataset using the show_batch method of the BioDataLoaders class. This method helps visualize the images and their corresponding labels, providing an overview of the dataset.
You can customize the display by modifying the following parameters: - max_n: The maximum number of images to display in the batch. By default, it shows all images in the batch. - nrows: The number of rows to use for displaying the images. This can be adjusted to control the layout of the images. - ncols: The number of columns to use for displaying the images. This can be adjusted to control the layout of the images. - figsize: The size of the figure used to display the images. This can be adjusted to make the images larger or smaller.
For example, you can set max_n=4 to display only 4 images from the batch, or set figsize=(10, 10) to increase the size of the displayed images.
This visualization step is useful for verifying that the images have been loaded and transformed correctly before proceeding with model training.
data.show_batch()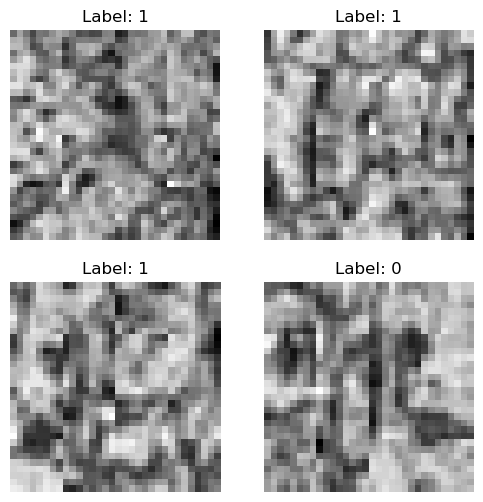
Load and train a 3D model
Train the Model
In the next cell, we will initialize and train a 3D model using the fastTrainer class. The model architecture used is SEResNet50, which is a 3D version of the SE-ResNet50 model. This model is well-suited for 3D image classification tasks.
We will use the following components: - SEResNet50: The model architecture with 3D spatial dimensions, 1 input channel, and 2 output classes. - CrossEntropyLossFlat: The loss function used for training the model. - BalancedAccuracy: The metric used to evaluate the model’s performance. - fastTrainer: A custom trainer class to handle the training process.
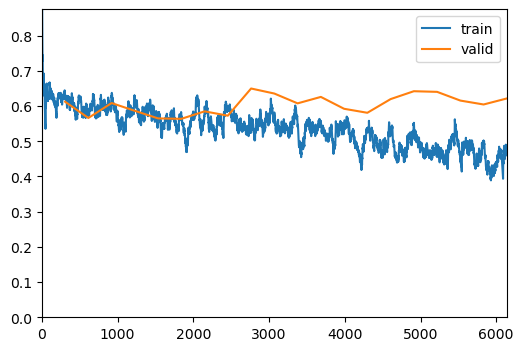
The trainer.fit(20) method will train the model for 20 epochs.
You can customize the training process by modifying the following parameters: - model: Change the model architecture to another 3D model, such as DenseNet169. - loss_fn: Use a different loss function, such as FocalLoss. - metrics: Add more metrics to evaluate the model, such as Precision or Recall. - show_summary: Set to False if you do not want to display the model summary. - find_lr: Set to False if you do not want to find the optimal learning rate.
For example, you can add more metrics to the metrics list to get a more comprehensive evaluation of the model’s performance.
You can customize the training process by modifying the following parameters: - epochs: Change the number of epochs to train the model for a different duration. For example, you can set it to 50 or 100 epochs. - lr: Adjust the learning rate to control the speed at which the model learns. A lower learning rate can lead to more stable training, while a higher learning rate can speed up the process but may cause instability. - callbacks: Add custom callbacks to monitor the training process, such as early stopping or learning rate schedulers.
For example, you can add an early stopping callback to stop training if the validation loss does not improve for a certain number of epochs. This can help prevent overfitting and save training time.
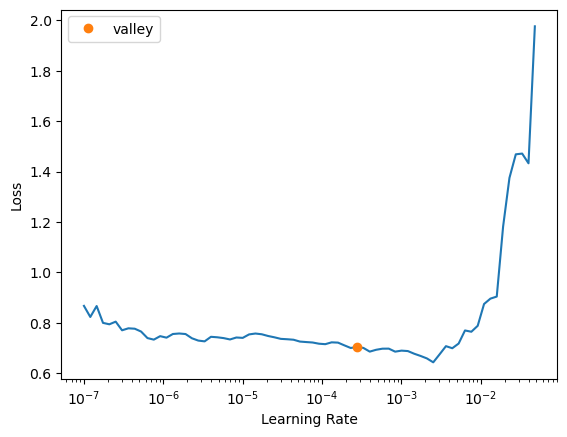
Additionally, you can experiment with different learning rates to find the optimal value for your dataset. The find_lr parameter in the fastTrainer class can help you automatically find a suitable learning rate.
By customizing these parameters, you can fine-tune the training process to achieve better performance and adapt the model to your specific dataset and requirements.
from monai.networks.nets import SEResNet50
from fastai.vision.all import BalancedAccuracy, CrossEntropyLossFlat
model = SEResNet50(spatial_dims=3, in_channels=1, num_classes=2)
loss = CrossEntropyLossFlat()
metrics = BalancedAccuracy()
trainer = fastTrainer(data, model, loss_fn=loss, metrics=metrics, show_summary=True, find_lr=True)SEResNet50 (Input shape: 4 x 1 x 32 x 32 x 32)
============================================================================
Layer (type) Output Shape Param # Trainable
============================================================================
4 x 64 x 16 x 16 x
Conv3d 21952 True
BatchNorm3d 128 True
ReLU
____________________________________________________________________________
4 x 64 x 8 x 8 x 8
MaxPool3d
Conv3d 4096 True
BatchNorm3d 128 True
ReLU
Conv3d 110592 True
BatchNorm3d 128 True
ReLU
____________________________________________________________________________
4 x 256 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 512 True
____________________________________________________________________________
4 x 256 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 16
Linear 4112 True
ReLU
____________________________________________________________________________
4 x 256
Linear 4352 True
Sigmoid
____________________________________________________________________________
4 x 256 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 64 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 128 True
ReLU
Conv3d 110592 True
BatchNorm3d 128 True
ReLU
____________________________________________________________________________
4 x 256 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 512 True
____________________________________________________________________________
4 x 256 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 16
Linear 4112 True
ReLU
____________________________________________________________________________
4 x 256
Linear 4352 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 64 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 128 True
ReLU
Conv3d 110592 True
BatchNorm3d 128 True
ReLU
____________________________________________________________________________
4 x 256 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 512 True
____________________________________________________________________________
4 x 256 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 16
Linear 4112 True
ReLU
____________________________________________________________________________
4 x 256
Linear 4352 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 128 x 4 x 4 x 4
Conv3d 32768 True
BatchNorm3d 256 True
ReLU
Conv3d 442368 True
BatchNorm3d 256 True
ReLU
____________________________________________________________________________
4 x 512 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 1024 True
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 32
Linear 16416 True
ReLU
____________________________________________________________________________
4 x 512
Linear 16896 True
Sigmoid
____________________________________________________________________________
4 x 512 x 4 x 4 x 4
Conv3d 131072 True
BatchNorm3d 1024 True
ReLU
____________________________________________________________________________
4 x 128 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 256 True
ReLU
Conv3d 442368 True
BatchNorm3d 256 True
ReLU
____________________________________________________________________________
4 x 512 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 1024 True
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 32
Linear 16416 True
ReLU
____________________________________________________________________________
4 x 512
Linear 16896 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 128 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 256 True
ReLU
Conv3d 442368 True
BatchNorm3d 256 True
ReLU
____________________________________________________________________________
4 x 512 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 1024 True
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 32
Linear 16416 True
ReLU
____________________________________________________________________________
4 x 512
Linear 16896 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 128 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 256 True
ReLU
Conv3d 442368 True
BatchNorm3d 256 True
ReLU
____________________________________________________________________________
4 x 512 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 1024 True
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 32
Linear 16416 True
ReLU
____________________________________________________________________________
4 x 512
Linear 16896 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 131072 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 524288 True
BatchNorm3d 2048 True
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 262144 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 262144 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 262144 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 262144 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 262144 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
Conv3d 524288 True
BatchNorm3d 1024 True
ReLU
Conv3d 7077888 True
BatchNorm3d 1024 True
ReLU
____________________________________________________________________________
4 x 2048 x 1 x 1 x
Conv3d 1048576 True
BatchNorm3d 4096 True
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 128
Linear 262272 True
ReLU
____________________________________________________________________________
4 x 2048
Linear 264192 True
Sigmoid
____________________________________________________________________________
4 x 2048 x 1 x 1 x
Conv3d 2097152 True
BatchNorm3d 4096 True
ReLU
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
Conv3d 1048576 True
BatchNorm3d 1024 True
ReLU
Conv3d 7077888 True
BatchNorm3d 1024 True
ReLU
____________________________________________________________________________
4 x 2048 x 1 x 1 x
Conv3d 1048576 True
BatchNorm3d 4096 True
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 128
Linear 262272 True
ReLU
____________________________________________________________________________
4 x 2048
Linear 264192 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
Conv3d 1048576 True
BatchNorm3d 1024 True
ReLU
Conv3d 7077888 True
BatchNorm3d 1024 True
ReLU
____________________________________________________________________________
4 x 2048 x 1 x 1 x
Conv3d 1048576 True
BatchNorm3d 4096 True
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 128
Linear 262272 True
ReLU
____________________________________________________________________________
4 x 2048
Linear 264192 True
Sigmoid
Identity
ReLU
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 2
Linear 4098 True
____________________________________________________________________________
Total params: 48,690,162
Total trainable params: 48,690,162
Total non-trainable params: 0
Optimizer used: <function Adam>
Loss function: FlattenedLoss of CrossEntropyLoss()
Callbacks:
- TrainEvalCallback
- CastToTensor
- Recorder
- ProgressCallback
- ShowGraphCallbackInferred learning rate: 4e-05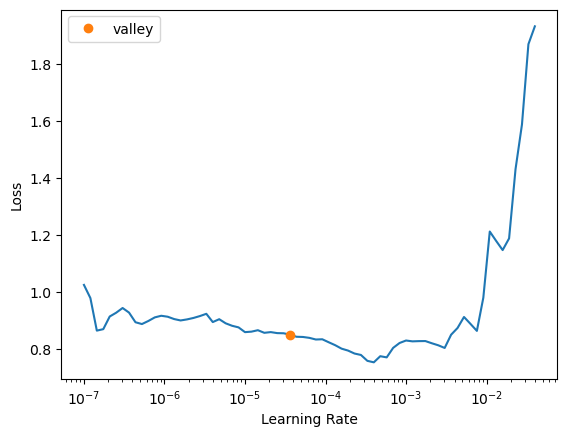
trainer.fit(20)| epoch | train_loss | valid_loss | balanced_accuracy_score | time |
|---|
Evaluate the Model on Validation Data
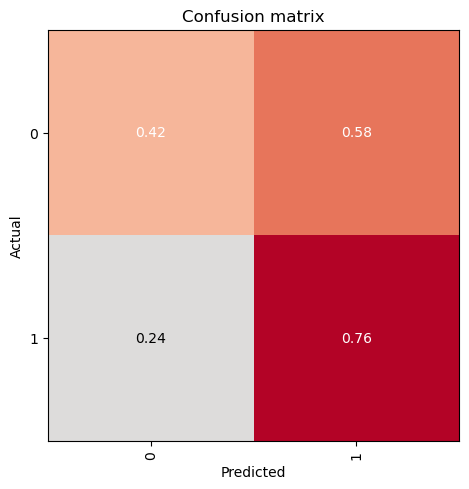
In this step, we will evaluate the trained model on the validation dataset using the evaluate_classification_model function. This function computes the specified metrics and provides insights into the model’s performance. Additionally, it can display the most confused classes to help identify areas for improvement.
- You can customize the
metricsparameter to include other evaluation metrics relevant to your task.- The
most_confused_nparameter specifies the number of most confused classes to display. Adjust this value to see more or fewer confused classes.- Set the
show_graphparameter toTrueto visualize the confusion matrix and other evaluation graphs.- Use this evaluation step to monitor the model’s performance and make necessary adjustments to the training process or data pipeline.
metrics = BalancedAccuracy()
evaluate_classification_model(trainer, most_confused_n=5, show_graph=False); precision recall f1-score support
0 0.39 0.42 0.40 48
1 0.78 0.76 0.77 129
accuracy 0.67 177
macro avg 0.58 0.59 0.59 177
weighted avg 0.67 0.67 0.67 177
Most Confused Classes:[('1', '0', 31), ('0', '1', 28)]| Value | |
|---|---|
| CrossEntropyLossFlat | |
| Mean | 0.616599 |
| Median | 0.561841 |
| Standard Deviation | 0.240548 |
| Min | 0.321632 |
| Max | 1.207438 |
| Q1 | 0.424600 |
| Q3 | 0.770084 |
Save the Trained Model
In the next cell, we will save the trained model to a file using the save method of the fastTrainer class. This step is important to preserve the trained model so that it can be loaded and used later without retraining.
You can customize the saving process by modifying the following parameters: - file_name: Change the name of the file to save the model with a different name. For example, you can set it to ‘final_model’ or ‘best_model’. - path: Specify a different path to save the model in a different directory. This is useful if you want to organize your saved models in a specific folder.
For example, you can set file_name='final_model' to save the model with the name ‘final_model.pth’, or set path='../models/' to save the model in the ‘models’ directory.
By customizing these parameters, you can ensure that the model is saved with a meaningful name and in an organized manner, making it easier to manage and retrieve the model for future use.
# trainer.save('tmp-model')Evaluate the Model on Test Data
Here, we will evaluate the performance of the trained model on the test dataset. This step is crucial to understand how well the model generalizes to unseen data.
We will use the data.test_dl method to create a DataLoader for the test dataset. The get_image_files function is used to retrieve the test images from the specified path. The with_labels=True parameter ensures that the test images are loaded with their corresponding labels.
After creating the test DataLoader, we will print the number of test images to verify that the dataset has been loaded correctly.
You can customize the evaluation process by modifying the following parameters: - test_path: Change the path to the test dataset if it is stored in a different location. - with_labels: Set to False if the test dataset does not have labels. This is useful for evaluating the model on unlabeled data. - batch_size: Adjust the batch size for the test DataLoader. A larger batch size can speed up the evaluation process but may require more memory.
By customizing these parameters, you can adapt the evaluation process to different datasets and requirements, ensuring that the model’s performance is accurately assessed.
test_data = data.test_dl(get_image_files(test_path), with_labels=True)
# print length of test dataset
print('test images:', len(test_data.items))test images: 352In the next cell, we will evaluate the performance of the trained model on the test dataset using the evaluate_classification_model function. This function will compute various evaluation metrics and display the results.
You can customize the evaluation process by modifying the following parameters: - show_graph: Set to True to display a graph of the evaluation metrics. This can help visualize the model’s performance. - show_results: Set to True to display the results of the evaluation, including the predicted labels and ground truth labels.
For example, you can set show_graph=True to visualize the evaluation metrics, or set show_results=True to see the detailed results of the evaluation.
By customizing these parameters, you can gain a deeper understanding of the model’s performance and identify areas for improvement. This step is crucial for fine-tuning the model and ensuring that it generalizes well to unseen data.
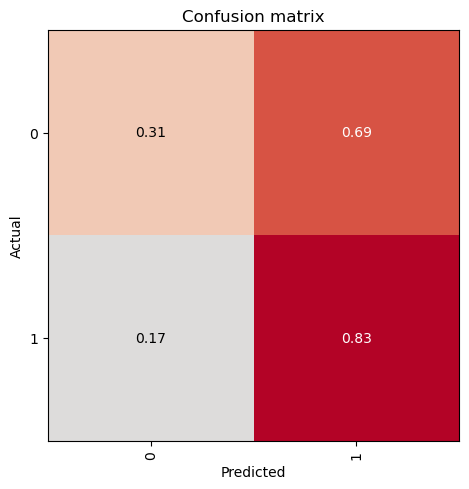
evaluate_classification_model(trainer, test_data, show_graph=False, show_results=False); precision recall f1-score support
0 0.40 0.31 0.35 95
1 0.76 0.83 0.79 257
accuracy 0.69 352
macro avg 0.58 0.57 0.57 352
weighted avg 0.66 0.69 0.67 352
Most Confused Classes:[('0', '1', 66), ('1', '0', 44)]| Value | |
|---|---|
| CrossEntropyLossFlat | |
| Mean | 0.613168 |
| Median | 0.525626 |
| Standard Deviation | 0.254610 |
| Min | 0.317651 |
| Max | 1.287738 |
| Q1 | 0.413106 |
| Q3 | 0.774548 |
Load the Model
In this step, we will load the previously trained model using the load method of the visionTrainer class. In this example, we will:
- Create a trainer instance and load the previously saved model.
- Fine tune the model a several epochs more.
- Evaluate the model with test data again.
model = SEResNet50(spatial_dims=3, in_channels=1, num_classes=2)
loss = CrossEntropyLossFlat()
metrics = BalancedAccuracy()
trainer2 = fastTrainer(data, model, loss_fn=loss, metrics=metrics, show_summary=True, find_lr=True)
# Load saved model
trainer2.load('tmp-model')
# Train several additional epochs
trainer2.fit_one_cycle(2, lr_max=4e-2)
# Evaluate the model on the test dataset
evaluate_classification_model(trainer2, test_data, show_graph=False);SEResNet50 (Input shape: 4 x 1 x 32 x 32 x 32)
============================================================================
Layer (type) Output Shape Param # Trainable
============================================================================
4 x 64 x 16 x 16 x
Conv3d 21952 True
BatchNorm3d 128 True
ReLU
____________________________________________________________________________
4 x 64 x 8 x 8 x 8
MaxPool3d
Conv3d 4096 True
BatchNorm3d 128 True
ReLU
Conv3d 110592 True
BatchNorm3d 128 True
ReLU
____________________________________________________________________________
4 x 256 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 512 True
____________________________________________________________________________
4 x 256 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 16
Linear 4112 True
ReLU
____________________________________________________________________________
4 x 256
Linear 4352 True
Sigmoid
____________________________________________________________________________
4 x 256 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 64 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 128 True
ReLU
Conv3d 110592 True
BatchNorm3d 128 True
ReLU
____________________________________________________________________________
4 x 256 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 512 True
____________________________________________________________________________
4 x 256 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 16
Linear 4112 True
ReLU
____________________________________________________________________________
4 x 256
Linear 4352 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 64 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 128 True
ReLU
Conv3d 110592 True
BatchNorm3d 128 True
ReLU
____________________________________________________________________________
4 x 256 x 8 x 8 x 8
Conv3d 16384 True
BatchNorm3d 512 True
____________________________________________________________________________
4 x 256 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 16
Linear 4112 True
ReLU
____________________________________________________________________________
4 x 256
Linear 4352 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 128 x 4 x 4 x 4
Conv3d 32768 True
BatchNorm3d 256 True
ReLU
Conv3d 442368 True
BatchNorm3d 256 True
ReLU
____________________________________________________________________________
4 x 512 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 1024 True
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 32
Linear 16416 True
ReLU
____________________________________________________________________________
4 x 512
Linear 16896 True
Sigmoid
____________________________________________________________________________
4 x 512 x 4 x 4 x 4
Conv3d 131072 True
BatchNorm3d 1024 True
ReLU
____________________________________________________________________________
4 x 128 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 256 True
ReLU
Conv3d 442368 True
BatchNorm3d 256 True
ReLU
____________________________________________________________________________
4 x 512 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 1024 True
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 32
Linear 16416 True
ReLU
____________________________________________________________________________
4 x 512
Linear 16896 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 128 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 256 True
ReLU
Conv3d 442368 True
BatchNorm3d 256 True
ReLU
____________________________________________________________________________
4 x 512 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 1024 True
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 32
Linear 16416 True
ReLU
____________________________________________________________________________
4 x 512
Linear 16896 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 128 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 256 True
ReLU
Conv3d 442368 True
BatchNorm3d 256 True
ReLU
____________________________________________________________________________
4 x 512 x 4 x 4 x 4
Conv3d 65536 True
BatchNorm3d 1024 True
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 32
Linear 16416 True
ReLU
____________________________________________________________________________
4 x 512
Linear 16896 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 131072 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 524288 True
BatchNorm3d 2048 True
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 262144 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 262144 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 262144 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 262144 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 256 x 2 x 2 x 2
Conv3d 262144 True
BatchNorm3d 512 True
ReLU
Conv3d 1769472 True
BatchNorm3d 512 True
ReLU
____________________________________________________________________________
4 x 1024 x 2 x 2 x
Conv3d 262144 True
BatchNorm3d 2048 True
____________________________________________________________________________
4 x 1024 x 1 x 1 x
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 64
Linear 65600 True
ReLU
____________________________________________________________________________
4 x 1024
Linear 66560 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
Conv3d 524288 True
BatchNorm3d 1024 True
ReLU
Conv3d 7077888 True
BatchNorm3d 1024 True
ReLU
____________________________________________________________________________
4 x 2048 x 1 x 1 x
Conv3d 1048576 True
BatchNorm3d 4096 True
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 128
Linear 262272 True
ReLU
____________________________________________________________________________
4 x 2048
Linear 264192 True
Sigmoid
____________________________________________________________________________
4 x 2048 x 1 x 1 x
Conv3d 2097152 True
BatchNorm3d 4096 True
ReLU
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
Conv3d 1048576 True
BatchNorm3d 1024 True
ReLU
Conv3d 7077888 True
BatchNorm3d 1024 True
ReLU
____________________________________________________________________________
4 x 2048 x 1 x 1 x
Conv3d 1048576 True
BatchNorm3d 4096 True
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 128
Linear 262272 True
ReLU
____________________________________________________________________________
4 x 2048
Linear 264192 True
Sigmoid
Identity
ReLU
____________________________________________________________________________
4 x 512 x 1 x 1 x 1
Conv3d 1048576 True
BatchNorm3d 1024 True
ReLU
Conv3d 7077888 True
BatchNorm3d 1024 True
ReLU
____________________________________________________________________________
4 x 2048 x 1 x 1 x
Conv3d 1048576 True
BatchNorm3d 4096 True
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 128
Linear 262272 True
ReLU
____________________________________________________________________________
4 x 2048
Linear 264192 True
Sigmoid
Identity
ReLU
AdaptiveAvgPool3d
____________________________________________________________________________
4 x 2
Linear 4098 True
____________________________________________________________________________
Total params: 48,690,162
Total trainable params: 48,690,162
Total non-trainable params: 0
Optimizer used: <function Adam>
Loss function: FlattenedLoss of CrossEntropyLoss()
Callbacks:
- TrainEvalCallback
- CastToTensor
- Recorder
- ProgressCallback
- ShowGraphCallbackInferred learning rate: 0.0003--------------------------------------------------------------------------- FileNotFoundError Traceback (most recent call last) Cell In[13], line 9 6 trainer2 = fastTrainer(data, model, loss_fn=loss, metrics=metrics, show_summary=True, find_lr=True) 8 # Load saved model ----> 9 trainer2.load('tmp-model') 11 # Train several additional epochs 12 trainer2.fit_one_cycle(2, lr_max=4e-2) File ~/anaconda3/envs/bioMONAI-env_linux/lib/python3.11/site-packages/fastai/learner.py:422, in load(self, file, device, **kwargs) 420 file = join_path_file(file, self.path/self.model_dir, ext='.pth') 421 distrib_barrier() --> 422 load_model(file, self.model, self.opt, device=device, **kwargs) 423 return self File ~/anaconda3/envs/bioMONAI-env_linux/lib/python3.11/site-packages/fastai/learner.py:53, in load_model(file, model, opt, with_opt, device, strict, **torch_load_kwargs) 51 if isinstance(device, int): device = torch.device('cuda', device) 52 elif device is None: device = 'cpu' ---> 53 state = torch.load(file, map_location=device, **torch_load_kwargs) 54 hasopt = set(state)=={'model', 'opt'} 55 model_state = state['model'] if hasopt else state File ~/anaconda3/envs/bioMONAI-env_linux/lib/python3.11/site-packages/torch/serialization.py:791, in load(f, map_location, pickle_module, weights_only, **pickle_load_args) 788 if 'encoding' not in pickle_load_args.keys(): 789 pickle_load_args['encoding'] = 'utf-8' --> 791 with _open_file_like(f, 'rb') as opened_file: 792 if _is_zipfile(opened_file): 793 # The zipfile reader is going to advance the current file position. 794 # If we want to actually tail call to torch.jit.load, we need to 795 # reset back to the original position. 796 orig_position = opened_file.tell() File ~/anaconda3/envs/bioMONAI-env_linux/lib/python3.11/site-packages/torch/serialization.py:271, in _open_file_like(name_or_buffer, mode) 269 def _open_file_like(name_or_buffer, mode): 270 if _is_path(name_or_buffer): --> 271 return _open_file(name_or_buffer, mode) 272 else: 273 if 'w' in mode: File ~/anaconda3/envs/bioMONAI-env_linux/lib/python3.11/site-packages/torch/serialization.py:252, in _open_file.__init__(self, name, mode) 251 def __init__(self, name, mode): --> 252 super().__init__(open(name, mode)) FileNotFoundError: [Errno 2] No such file or directory: '../_data/medmnist_data/synapsemnist3d/models/tmp-model.pth'